Lecture
PL/R (Programming Language/Relational) is a procedural language extension for the PostgreSQL database system. It allows you to write functions and procedures using the R programming language within the context of a PostgreSQL database. PL/R can be used to perform complex data analysis and statistical computations directly within the database, leveraging the power of R's statistical and analytical capabilities.
Here are some key features and capabilities of PL/R:
R Integration: PL/R allows you to write database functions and procedures using the R programming language. This enables you to perform data analysis, modeling, and statistical computations using R's extensive libraries and functions.
Data Analysis: With PL/R, you can create stored procedures that perform various types of data analysis directly within the database. This can include tasks such as regression analysis, clustering, data visualization, and more.
Statistical Functions: You can define custom statistical functions in R and call them from SQL queries, allowing you to compute statistical measures and summaries on your data.
Data Transformation: PL/R can be used to preprocess and transform data stored in the database. This includes cleaning, filtering, and reshaping data before performing analysis.
Parallel Processing: PL/R can take advantage of parallel processing capabilities offered by PostgreSQL, allowing you to distribute computations across multiple processors or cores.
Scalability: Since PL/R operates within the PostgreSQL database, it benefits from PostgreSQL's scalability features, such as support for large datasets, indexing, and query optimization.
Integration with SQL: PL/R functions can be seamlessly integrated with SQL queries. This means you can use SQL to query and manipulate your data and call PL/R functions for more complex analysis tasks.
Custom Aggregates: PL/R allows you to create custom aggregate functions that can be used in SQL queries. These aggregates can perform specialized statistical operations on groups of data.
Machine Learning: You can leverage R's machine learning libraries to build and apply predictive models within the database.
Keep in mind that while PL/R provides powerful capabilities for data analysis and statistical computations, it might require a good understanding of both SQL and R to effectively utilize its features. It's important to carefully design your functions and procedures to optimize performance and leverage the strengths of both PostgreSQL and R.
Please note that my knowledge is based on information available up to September 2021, and there might have been developments or changes since then. If you're interested in using PL/R, I recommend referring to the official PostgreSQL documentation and resources for the most up-to-date information.
func_name <- function(myarg1 [,myarg2...]) { function body referencing myarg1 [, myarg2 ...] }
CREATE OR REPLACE FUNCTION func_name(arg-type1 [, arg-type2 ...]) RETURNS return-type AS $$ function body referencing arg1 [, arg2 ...] $$ LANGUAGE 'plr'; CREATE OR REPLACE FUNCTION func_name(myarg1 arg-type1 [, myarg2 arg-type2 ...]) RETURNS return-type AS $$ function body referencing myarg1 [, myarg2 ...] $$ LANGUAGE 'plr';
CREATE EXTENSION plr; CREATE OR REPLACE FUNCTION test_dtup(OUT f1 text, OUT f2 int) RETURNS SETOF record AS $$ data.frame(letters[1:3],1:3) $$ LANGUAGE 'plr'; SELECT * FROM test_dtup(); f1 | f2 ----+---- a | 1 b | 2 c | 3 (3 rows)
dbDriver(character dvr_name) dbConnect(DBIDriver drv, character user, character password, character host, character dbname, character port, character tty, character options) dbSendQuery(DBIConnection conn, character sql) fetch(DBIResult rs, integer num_rows) dbClearResult (DBIResult rs) dbGetQuery(DBIConnection conn, character sql) dbReadTable(DBIConnection conn, character name) dbDisconnect(DBIConnection conn) dbUnloadDriver(DBIDriver drv)
tsp_tour_length<-function() { require(TSP) require(fields) require(RPostgreSQL) drv <- dbDriver("PostgreSQL") conn <- dbConnect(drv, user="postgres", dbname="plr", host="localhost") sql.str <- "select id, st_x(location) as x, st_y(location) as y, location from stands" waypts <- dbGetQuery(conn, sql.str) dist.matrix <- rdist.earth(waypts[,2:3], R=3949.0) rtsp <- TSP(dist.matrix) soln <- solve_TSP(rtsp) dbDisconnect(conn) dbUnloadDriver(drv) return(attributes(soln)$tour_length) }
CREATE OR REPLACE FUNCTION tsp_tour_length() RETURNS float8 AS $$ require(TSP) require(fields) require(RPostgreSQL) drv <- dbDriver("PostgreSQL") conn <- dbConnect(drv, user="postgres", dbname="plr", host="localhost") sql.str <- "select id, st_x(location) as x, st_y(location) as y, location from stands" waypts <- dbGetQuery(conn, sql.str) dist.matrix <- rdist.earth(waypts[,2:3], R=3949.0) rtsp <- TSP(dist.matrix) soln <- solve_TSP(rtsp) dbDisconnect(conn) dbUnloadDriver(drv) return(attributes(soln)$tour_length) $$ LANGUAGE 'plr' STRICT;
tsp_tour_length() [1] 2804.581
SELECT tsp_tour_length(); tsp_tour_length ------------------ 2804.58129355858 (1 row)
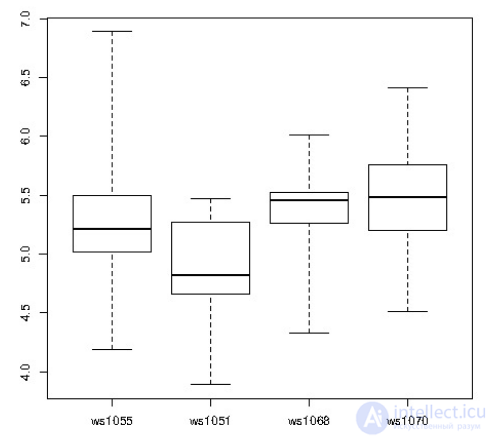
CREATE OR REPLACE FUNCTION r_quartile(ANYARRAY) RETURNS ANYARRAY AS $$ quantile(arg1, probs = seq(0, 1, 0.25), names = FALSE) $$ LANGUAGE 'plr'; CREATE AGGREGATE quartile (ANYELEMENT) ( sfunc = array_append, stype = ANYARRAY, finalfunc = r_quantile, initcond = '{}' ); SELECT workstation, quartile(id_val) FROM sample_numeric_data WHERE ia_id = 'G121XB8A' GROUP BY workstation; workstation | quantile -------------+--------------------------------- 1055 | {4.19,5.02,5.21,5.5,6.89} 1051 | {3.89,4.66,4.825,5.2675,5.47} 1068 | {4.33,5.2625,5.455,5.5275,6.01} 1070 | {4.51,5.1975,5.485,5.7575,6.41} (4 rows)
CREATE TABLE test_data ( fyear integer, firm float8, eps float8 ); INSERT INTO test_data SELECT (bf + 1) % 10 + 2000 AS fyear, floor((b.f+1)/10) + 50 AS firm, f::float8/100 + random()/10 AS eps FROM generate_series(-500,499,1) b(f); CREATE OR REPLACE FUNCTION r_regr_slope(float8, float8) RETURNS float8 AS $BODY$ slope <- NA y <- farg1 x <- farg2 if (fnumrows==9) try (slope <- lm(y ~ x)$coefficients[2]) return(slope) $BODY$ LANGUAGE plr WINDOW;
SELECT *, r_regr_slope(eps, lag_eps) OVER w AS slope_R FROM ( SELECT firm AS f, fyear AS fyr, eps, lag(eps) OVER (PARTITION BY firm ORDER BY firm, fyear) AS lag_eps FROM test_data ) AS a WHERE eps IS NOT NULL WINDOW w AS (PARTITION BY firm ORDER BY firm, fyear ROWS 8 PRECEDING); f | fyr | eps | lag_eps | slope_r ---+------+-------------------+-------------------+------------------- 1 | 1991 | -4.99563754182309 | | 1 | 1992 | -4.96425441872329 | -4.99563754182309 | 1 | 1993 | -4.96906093481928 | -4.96425441872329 | 1 | 1994 | -4.92376988714561 | -4.96906093481928 | 1 | 1995 | -4.95884547665715 | -4.92376988714561 | 1 | 1996 | -4.93236254784279 | -4.95884547665715 | 1 | 1997 | -4.90775520844385 | -4.93236254784279 | 1 | 1998 | -4.92082695348188 | -4.90775520844385 | 1 | 1999 | -4.84991340579465 | -4.92082695348188 | 0.691850614092383 1 | 2000 | -4.86000917562284 | -4.84991340579465 | 0.700526929134053
install.packages(c('xts','Defaults','quantmod','cairoDevice','RGtk2'))
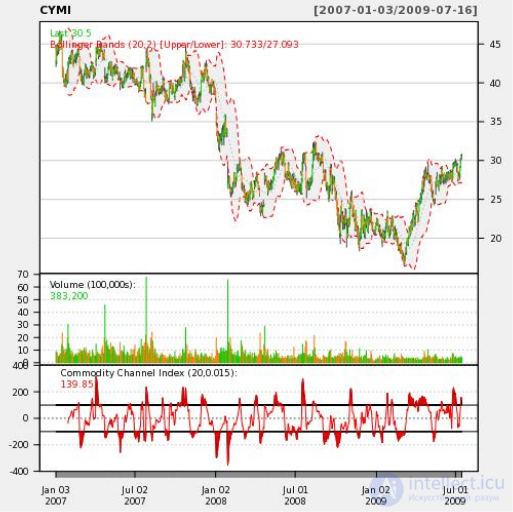
CREATE OR REPLACE FUNCTION plot_stock_data(sym text) RETURNS bytea AS $$ library(quantmod) library(cairoDevice) library(RGtk2) pixmap <- gdkPixmapNew(w=500, h=500, depth=24) asCairoDevice(pixmap) getSymbols(c(sym)) chartSeries(get(sym), name=sym, theme="white", TA="addVo();addBBands();addCCI()") plot_pixbuf <- gdkPixbufGetFromDrawable(NULL, pixmap, pixmap$getColormap(),0, 0, 0, 0, 500, 500) buffer <- gdkPixbufSaveToBufferv(plot_pixbuf, "jpeg", character(0), character(0))$buffer return(buffer) $$ LANGUAGE plr;
Xvfb :1 -screen 0 1024x768x24 export DISPLAY=:1.0
CREATE TABLE open_emv_cost(value float8, district int); COPY open_emv_cost FROM 'open-emv.cost.csv' WITH delimiter ',';
CREATE TYPE benford_t AS ( actual_mean float8, n int, expected_mean float8, distorion float8, z float8 ); CREATE OR REPLACE FUNCTION benford(numarr float8[]) RETURNS benford_t AS $$ xcoll <- function(x) { return ((10 * x) / (10 ^ (trunc(log10(x))))) } numarr <- numarr[numarr >= 10] numarr <- xcoll(numarr) actual_mean <- mean(numarr) n <- length(numarr) expected_mean <- (90 / (n * (10 ^ (1/n) - 1))) distorion <- ((actual_mean - expected_mean) / expected_mean) z <- (distorion / sd(numarr)) retval <- data.frame(actual_mean,n,expected_mean,distorion,z) return(retval) $$ LANGUAGE plr;
SELECT * FROM benford(array(SELECT value FROM open_emv_cost)); -[ RECORD 1 ]-+---------------------- actual_mean | 38.1936561918275 n | 240 expected_mean | 38.8993031865999 distorion | -0.0181403505195804 z | -0.000984036908080443
CREATE TABLE stands ( id serial primary key, strata integer not null, initage integer ); SELECT AddGeometryColumn('','stands','boundary','4326','MULTIPOLYGON',2); CREATE INDEX "stands_boundary_gist" ON "stands" USING gist ("boundary" gist_geometry_ops); SELECT AddGeometryColumn('','stands','location','4326','POINT',2); CREATE INDEX "stands_location_gist" ON "stands" USING gist ("location" gist_geometry_ops); INSERT INTO stands (id,strata,initage,boundary,location) VALUES ( 1,1,1, GeometryFromText( 'MULTIPOLYGON(((59.250000 65.000000,55.000000 65.000000,55.000000 51.750000, 60.735294 53.470588, 62.875000 57.750000, 59.250000 65.000000 )))', 4326 ), GeometryFromText('POINT( 61.000000 59.000000 )', 4326) ), ( 2,2,1, GeometryFromText( 'MULTIPOLYGON(((67.000000 65.000000,59.250000 65.000000,62.875000 57.750000, 67.000000 60.500000, 67.000000 65.000000 )))', 4326 ), GeometryFromText('POINT( 63.000000 60.000000 )', 4326 ) ), ( 3,3,1, GeometryFromText( 'MULTIPOLYGON(((67.045455 52.681818,60.735294 53.470588,55.000000 51.750000, 55.000000 45.000000, 65.125000 45.000000, 67.045455 52.681818 )))', 4326 ), GeometryFromText('POINT( 64.000000 49.000000 )', 4326 ) ) ;
INSERT INTO stands (id,strata,initage,boundary,location) VALUES ( 4,4,1, GeometryFromText( 'MULTIPOLYGON(((71.500000 53.500000,70.357143 53.785714,67.045455 52.681818, 65.125000 45.000000, 71.500000 45.000000, 71.500000 53.500000 )))', 4326 ), GeometryFromText('POINT( 68.000000 48.000000 )', 4326) ), ( 5,5,1, GeometryFromText( 'MULTIPOLYGON(((69.750000 65.000000,67.000000 65.000000,67.000000 60.500000, 70.357143 53.785714, 71.500000 53.500000, 74.928571 54.642857, 69.750000 65.000000 )))', 4326 ), GeometryFromText('POINT( 71.000000 60.000000 )', 4326) ), ( 6,6,1, GeometryFromText( 'MULTIPOLYGON(((80.000000 65.000000,69.750000 65.000000,74.928571 54.642857, 80.000000 55.423077, 80.000000 65.000000 )))', 4326 ), GeometryFromText('POINT( 73.000000 61.000000 )', 4326) ), ( 7,7,1, GeometryFromText( 'MULTIPOLYGON(((80.000000 55.423077,74.928571 54.642857,71.500000 53.500000, 71.500000 45.000000, 80.000000 45.000000, 80.000000 55.423077 )))', 4326 ), GeometryFromText('POINT( 75.000000 48.000000 )', 4326) ), ( 8,8,1, GeometryFromText( 'MULTIPOLYGON(((67.000000 60.500000,62.875000 57.750000,60.735294 53.470588, 67.045455 52.681818, 70.357143 53.785714, 67.000000 60.500000 )))', 4326 ), GeometryFromText('POINT( 65.000000 57.000000 )', 4326) ) ;
DROP TABLE IF EXISTS events CASCADE; CREATE TABLE events ( seqid int not null primary key, -- visit sequence # plotid int, -- original plot id bearing real, -- bearing to next waypoint distance real, -- distance to next waypoint velocity real, -- velocity of travel, in nm/hr traveltime real, -- travel time to next event loitertime real, -- how long to hang out totaltraveldist real, -- cummulative distance totaltraveltime real -- cummulaative time ); SELECT AddGeometryColumn('','events','location','4326','POINT',2); CREATE INDEX "events_location_gist" ON "events" USING gist ("location" gist_geometry_ops); CREATE TABLE plr_modules ( modseq int4 primary key, modsrc text );
CREATE OR REPLACE FUNCTION solve_tsp(makemap bool, mapname text) RETURNS SETOF events AS $$ require(TSP) require(fields) sql.str <- "select id, st_x(location) as x, st_y(location) as y, location from stands;" waypts <- pg.spi.exec(sql.str) dist.matrix <- rdist.earth(waypts[,2:3], R=3949.0) rtsp <- TSP(dist.matrix) soln <- solve_TSP(rtsp) tour <- as.vector(soln) pg.thrownotice( paste("tour.dist=", attributes(soln)$tour_length)) route <- make.route(tour, waypts, dist.matrix) if (makemap) { make.map(tour, waypts, mapname) } return(route) $$ LANGUAGE 'plr' STRICT;
INSERT INTO plr_modules VALUES ( 0, $$ make.route <-function(tour, waypts, dist.matrix) { velocity <- 500.0 starts <- tour[1:(length(tour))-1] stops <- tour[2:(length(tour))] dist.vect <- diag( as.matrix( dist.matrix )[starts,stops] ) last.leg <- as.matrix( dist.matrix )[tour[length(tour)],tour[1]] dist.vect <- c(dist.vect, last.leg ) delta.x <- diff( waypts[tour,]$x ) delta.y <- diff( waypts[tour,]$y ) bearings <- atan( delta.x/delta.y ) * 180 / pi bearings <- c(bearings,0) for( i in 1:(length(tour)-1) ) { if( delta.x[i] > 0.0 && delta.y[i] > 0.0 ) bearings[i] <- bearings[i] if( delta.x[i] > 0.0 && delta.y[i] < 0.0 ) bearings[i] <- 180.0 + bearings[i] if( delta.x[i] < 0.0 && delta.y[i] > 0.0 ) bearings[i] <- 360.0 + bearings[i] if( delta.x[i] < 0.0 && delta.y[i] < 0.0 ) bearings[i] <- 180 + bearings[i] } route <- data.frame(seq=1:length(tour), ptid=tour, bearing=bearings, dist.vect=dist.vect, velocity=velocity, travel.time=dist.vect/velocity, loiter.time=0.5) route$total.travel.dist <- cumsum(route$dist.vect) route$total.travel.time <- cumsum(route$travel.time+route$loiter.time) route$location <- waypts[tour,]$location return(route)}$$ );
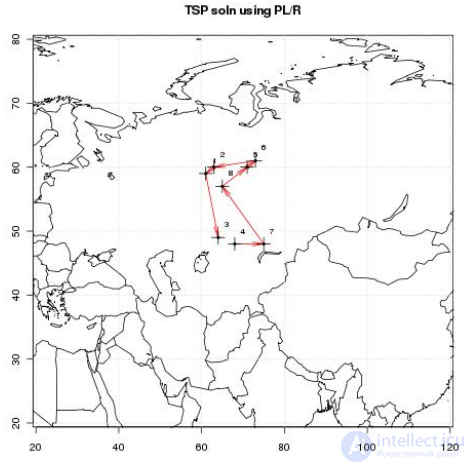
INSERT INTO plr_modules VALUES ( 1, $$make.map <-function(tour, waypts, mapname) { require(maps) jpeg(file=mapname, width = 480, height = 480, pointsize = 10, quality = 75) map('world2', xlim = c(20, 120), ylim=c(20,80) ) map.axes() grid() arrows( waypts[tour[1:(length(tour)-1)],]$x, waypts[tour[1:(length(tour)-1)],]$y, waypts[tour[2:(length(tour))],]$x, waypts[tour[2:(length(tour))],]$y, angle=10, lwd=1, length=.15, col="red" ) points( waypts$x, waypts$y, pch=3, cex=2 ) points( waypts$x, waypts$y, pch=20, cex=0.8 ) text( waypts$x+2, waypts$y+2, as.character( waypts$id ), cex=0.8 ) title( "TSP soln using PL/R" ) dev.off() }$$ );
-- only needed if INSERT INTO plr_modules was in same session SELECT reload_plr_modules(); SELECT seqid, plotid, bearing, distance, velocity, traveltime, loitertime, totaltraveldist FROM solve_tsp(true, 'tsp.jpg'); NOTICE: tour.dist= 2804.58129355858 seqid | plotid | bearing | distance | velocity | traveltime | loitertime | totaltraveldist -------+--------+---------+----------+----------+------------+------------+----------------- 1 | 8 | 131.987 | 747.219 | 500 | 1.49444 | 0.5 | 747.219 2 | 7 | -90 | 322.719 | 500 | 0.645437 | 0.5 | 1069.94 3 | 4 | 284.036 | 195.219 | 500 | 0.390438 | 0.5 | 1265.16 4 | 3 | 343.301 | 699.683 | 500 | 1.39937 | 0.5 | 1964.84 5 | 1 | 63.4349 | 98.2015 | 500 | 0.196403 | 0.5 | 2063.04 6 | 2 | 84.2894 | 345.957 | 500 | 0.691915 | 0.5 | 2409 7 | 6 | 243.435 | 96.7281 | 500 | 0.193456 | 0.5 | 2505.73 8 | 5 | 0 | 298.855 | 500 | 0.59771 | 0.5 | 2804.58 (8 rows)
\x SELECT * FROM solve_tsp(true, 'tsp.jpg'); NOTICE: tour.dist= 2804.58129355858 -[ RECORD 1 ]---+--------------------------------------------------- seqid | 1 plotid | 3 bearing | 104.036 distance | 195.219 velocity | 500 traveltime | 0.390438 loitertime | 0.5 totaltraveldist | 195.219 totaltraveltime | 0.890438 location | 0101000020E610000000000000000050400000000000804840 -[ RECORD 2 ]---+--------------------------------------------------- [...]
DROP TABLE IF EXISTS test_ts; CREATE TABLE test_ts ( dataid bigint NOT NULL PRIMARY KEY, data double precision[] ); CREATE OR REPLACE FUNCTION load_test(int) RETURNS text AS $$ DECLARE i int; arr text; sql text; BEGIN arr := pg_read_file('array-data.csv', 0, 500000); FOR i IN 1..$1 LOOP sql := $i$INSERT INTO test_ts(dataid,data) VALUES ($i$ || i || $i$,'{$i$ || arr || $i$}')$i$; EXECUTE sql; END LOOP; RETURN 'OK'; END; $$ LANGUAGE plpgsql; SELECT load_test(1000); load_test ----------- OK (1 row) Time: 37336.539 ms
DROP TABLE IF EXISTS test_ts_obj; CREATE TABLE test_ts_obj ( dataid serial PRIMARY KEY, data bytea ); CREATE OR REPLACE FUNCTION make_r_object(fname text) RETURNS bytea AS $$ myvar<-scan(fname,sep=",") return(myvar); $$ LANGUAGE 'plr' IMMUTABLE; INSERT INTO test_ts_obj (data) SELECT make_r_object('array-data.csv') FROM generate_series(1,1000); INSERT 0 1000 Time: 12166.137 ms
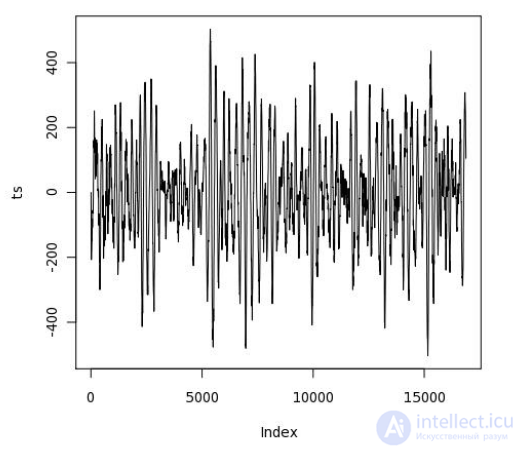
CREATE OR REPLACE FUNCTION plot_ts(ts double precision[]) RETURNS bytea AS $$ library(quantmod) library(cairoDevice) library(RGtk2) pixmap <- gdkPixmapNew(w=500, h=500, depth=24) asCairoDevice(pixmap) plot(ts,type="l") plot_pixbuf <- gdkPixbufGetFromDrawable( NULL, pixmap, pixmap$getColormap(), 0, 0, 0, 0, 500, 500 ) buffer <- gdkPixbufSaveToBufferv( plot_pixbuf, "jpeg", character(0), character(0) )$buffer return(buffer) $$ LANGUAGE 'plr' IMMUTABLE; SELECT plr_get_raw(plot_ts(data)) FROM test_ts WHERE dataid = 42;
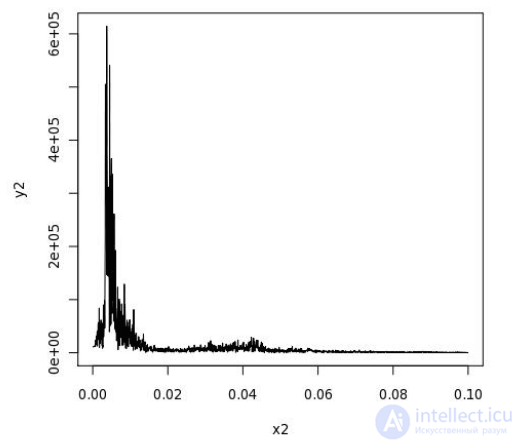
CREATE OR REPLACE FUNCTION plot_fftps(ts bytea) RETURNS bytea AS $$ library(quantmod) library(cairoDevice) library(RGtk2) fourier<-fft(ts) magnitude<-Mod(fourier) y2 <- magnitude[1:(length(magnitude)/10)] x2 <- 1:length(y2)/length(magnitude) mydf <- data.frame(x2,y2) pixmap <- gdkPixmapNew(w=500, h=500, depth=24) asCairoDevice(pixmap) plot(mydf,type="l") plot_pixbuf <- gdkPixbufGetFromDrawable( NULL, pixmap, pixmap$getColormap(), 0, 0, 0, 0, 500, 500 ) buffer <- gdkPixbufSaveToBufferv( plot_pixbuf, "jpeg", character(0), character(0) )$buffer return(buffer) $$ LANGUAGE 'plr' IMMUTABLE; SELECT plr_get_raw(plot_fftps(data)) FROM test_ts_obj WHERE dataid = 42;
Comments
To leave a comment
Databases, knowledge and data warehousing. Big data, DBMS and SQL and noSQL
Terms: Databases, knowledge and data warehousing. Big data, DBMS and SQL and noSQL